Our Motto is: “Research Never Stops”
Our research interests include biometrics, pattern recognition, image processing, and artificial intelligence.
Advanced Hand-based Biometrics: Recognition Algorithms, Systems, and Security Vulnerabilities

Biometric authentication based on hand features, including palmprint, finger knuckle print, dorsal hand veins, etc., offers a balance of high security and user convenience. Our research encompasses all aspects of this domain, addressing real-world challenges such as uncontrolled lighting, pose variations, and complex feature matching. To overcome these limitations, our team has pioneered a series of deep learning and representation-based frameworks. We have developed Deep Discriminative Representation for generic palmprint recognition (Pattern Recognition 2020) and Discriminant Direction Binary Descriptors to capture intrinsic features and coordinate-aware contrasts (IEEE TIP 2019). To further enhance feature robustness, we introduced Salient and Discriminative Descriptors for extraction and identification (IEEE TNNLS 2020), alongside Joint Constrained Least-Square Regression integrated with deep convolutional features (IEEE TSMC: Systems 2020). Furthermore, our work extends to finger vein and knuckle recognition using robust sparse least square regression (IEEE TIFS 2024). Addressing the critical security vulnerabilities of biometric systems, we have also investigated GAN-based data poisoning backdoor attacks and generative methods, exposing weaknesses in recognition CNNs to develop more resilient countermeasures (ICME 2025). These innovations, summarized in our monographs Advanced Hand-based Biometrics (Springer 2026) and Advanced Palmprint Authentication (Springer 2025), significantly enhance the accuracy and reliability of contactless and mobile biometric systems.
Medical Biometrics: Non-invasive Diagnosis via Intelligent Imaging
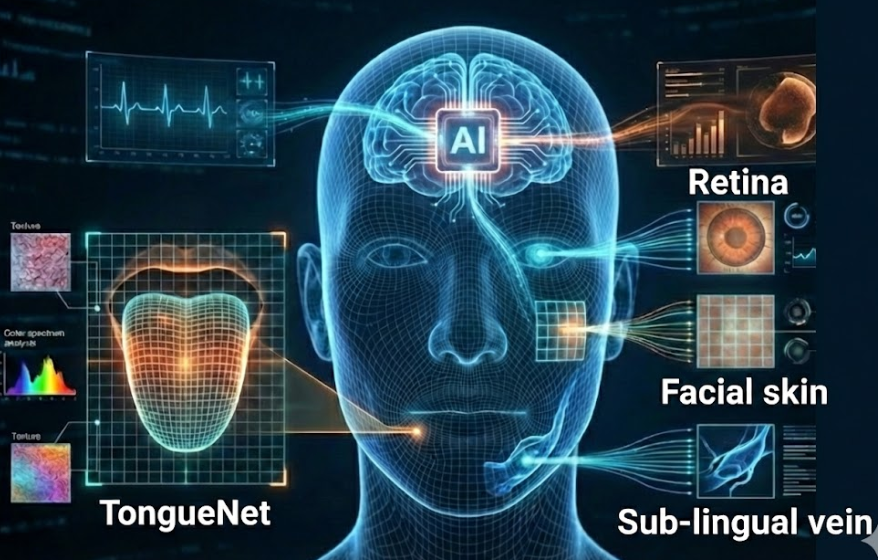
The integration of artificial intelligence with traditional medical diagnosis provides a pathway to non-invasive, objective, and efficient health monitoring. Traditional diagnostic methods, such as tongue and facial diagnosis, often lack standardization and quantitative analysis, while Western medicine can be invasive and time-consuming. Our research bridges this gap by developing computerized diagnostic systems. We have constructed advanced frameworks like TongueNet for precise tongue segmentation and feature extraction, covering color, texture, and geometry analysis (Applied Sciences 2019). Beyond tongue imaging, we utilize facial block color and texture features to detect chronic diseases such as diabetes mellitus and non-proliferative diabetic retinopathy (IEEE TBE 2013). To further enhance diagnostic precision, we developed an automatic multi-view disease detection system via Collective Deep Region-based Feature Representation, which fuses physiological information from the face, tongue, and sub-lingual veins (Future Generation Computer Systems 2021). By synthesizing these physiological features with sparse representation classifiers and deep neural networks, our work, highlighted in Advanced Medical Biometrics (World Scientific 2025), Facial Multi-characteristics and Applications (World Scientific 2018) and Tongue Image Analysis (Springer 2017), establishes a scientific foundation for computational medicine, enabling early detection and personalized healthcare monitoring.
Pattern Recognition: Robust Feature Learning and Representation

Fundamental pattern recognition algorithms are the engine driving high-performance AI systems, yet they often struggle with issues like high dimensionality, data noise, and small sample sizes. Our team focuses on developing mathematically rigorous and computationally efficient learning models. We have advanced the field of subspace learning by providing a definitive primer on Linear Discriminant Analysis (LDA) (Nature Reviews Methods Primers 2024) and developing robust variants such as latent linear discriminant analysis (Pattern Recognition 2024). We also pioneered Collaborative Representation and Sparse Representation based classifiers to handle robust visual classification tasks (IEEE TIP 2022, IEEE TIFS 2021, IEEE TMM 2021, IEEE TCYB 2020, IEEE TNNLS 2018, IEEE TBE 2014, Information Sciences 2012). Recent innovations include sophisticated Incomplete Multi-View Clustering frameworks. Specifically, we developed DIMC-net (ACM Multimedia 2020) and tensorized graph completion methods to effectively handle missing views in complex data (AAAI 2023, ACM Multimedia 2023). These contributions, thoroughly examined in our monograph Information Fusion: Machine Learning Methods (Springer 2022), provide the theoretical backbone for handling complex multimodal data, ensuring robustness and generalization across various visual understanding tasks.
Image Processing: Denoising, Restoration, and Enhancement via Generative AI

High-fidelity imaging is a prerequisite for accurate analysis, particularly in complex environments where data is corrupted by noise, blur, or low resolution. Our research addresses these inverse problems by leveraging cutting-edge generative AI and attention mechanisms. To achieve state-of-the-art performance in image denoising, we have developed QFormer (IJCAI 2024) and PID controller-guided attention networks (IEEE TNNLS 2021). For super-resolution, we proposed the Cosine Network (IEEE TIP 2025) and Tree-guided CNNs (IEEE TCE 2025), which effectively recover high-frequency details from low-resolution inputs. Furthermore, to enhance generalizability across diverse datasets and address the complexities of different camera sensors and in-camera signal processing pipelines, we introduced Dual Meta Attention (IEEE TCYB 2022) and Generative Adaptive Convolutions (AAAI 2022) for robust real-world photograph denoising. These technologies not only enhance visual quality, but also significantly improve the downstream performance of recognition and diagnostic systems.
Computational Biology: Phenotypic Learning and Virtual Cell Modeling

Understanding cellular phenotypes and predicting their responses to drug perturbations are pivotal challenges in modern computational biology. Traditional high-throughput screening often fails to capture subtle, high-dimensional morphological changes essential for accurate drug characterization. To bridge this gap, our research pioneers the intersection of AI and biotechnology. We developed PhenoProfiler (Nature Communications 2025), a flagship framework that advances phenotypic learning for image-based drug discovery, enabling the precise quantification of drug effects. Building on this foundation, we are advancing towards multimodal virtual cell models that integrate diverse data sources to simulate dynamic cellular behaviors under physiological and pathological conditions. By synergizing generative AI with large-scale phenotypic data, we aim to unravel the molecular mechanisms driving cell fate and optimize the drug development pipeline, marking a transformative step towards AI-driven precision medicine.
